 Embryo series courtesy of Einhard Schierenberg
Embryo series courtesy of Einhard SchierenbergTable of Contents
Abstract
Ubiquitin is a highly conserved 76 amino acid polypeptide, which is covalently attached to target proteins to signal their degradation by the 26S proteasome or to modify their function or localization. Regulated protein degradation, which is associated with many dynamic cellular processes, occurs predominantly via the ubiquitin-proteasome system. Ubiquitin is conjugated to target proteins through the sequential actions of a ubiquitin-activating enzyme, ubiquitin-conjugating enzymes, and ubiquitin-protein ligases. The nematode Caenorhabditis elegans has one ubiquitin-activating enzyme, twenty putative ubiquitin-conjugating enzymes, and potentially hundreds of ubiquitin-protein ligases. Research in C. elegans has focused on the cellular functions of ubiquitin pathway components in the context of organismal development. A combination of forward genetics, reverse genetics, and genome-wide RNAi screens has provided information on the loss-of-function phenotypes for the majority of C. elegans ubiquitin pathway components. Additionally, detailed analysis of several classes of ubiquitin-protein ligases has led to the identification of their substrates and the molecular pathways that they regulate. This review presents a comprehensive overview of ubiquitin-mediated pathways in C. elegans with a description of the known components and their identified molecular, cellular, and developmental functions.
Copyright: © 2005 Edward T. Kipreos. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Ubiquitin (Ub) is a ubiquitously expressed and highly conserved 76 amino acid polypeptide (Hershko and Ciechanover, 1998; Figure 1A). The covalent tandem attachment of multiple Ub to a target protein to form poly-ubiquitin chains can mark the protein for degradation by the 26S proteasome. Ub is covalently attached to substrate proteins by the concerted actions of three classes of enzymes (Hershko and Ciechanover, 1998). A ubiquitin-activating enzyme (E1) uses one ATP molecule to bind Ub via a thiolester linkage. The activated Ub is transferred to a ubiquitin-conjugating enzyme (E2), also via a thiolester linkage. The E2 is brought to the substrate by binding a ubiquitin-protein ligase (E3) that binds both the E2 and the substrate. Once bound to an E3, the E2 either directly transfers Ub to the substrate or transfers the Ub through a thiolester linkage to the E3, which then transfers the Ub to the substrate. Multiple rounds of E2 interactions with substrate-bound E3 are required to produce a poly-Ub chain on the substrate. In a few cases, the E2-E3 combination is not capable of adding more than a few Ub, and in this situation a ubiquitin chain assembly factor (E4) is required for the conjugation of additional Ub to form a poly-Ub chain (Koegl et al., 1999).

Figure 1. A) Alignment of individual ubiquitin polypeptides from C. elegans (C.e.), H. sapiens (H.s.), and S. cerevisiae (S.c.). Differences between Ub residues are boxed. Note that C. elegans and humans only differ at one position relative to each other and at three positions relative to budding yeast. The locations of the seven lysines in Ub are marked with the residue numbers provided above the alignment. B) Translation of the C. elegans ubq-1 polyubiquitin gene. The amino acid sequence is presented at 76 amino acids per line so that individual ubiquitin repeats are aligned. Note that repeat 6 has a different amino acid at position 9 of the repeat (yellow box, isoleucine rather than threonine). There are two amino acids after the final ubiquitin repeat.
Ub is conjugated to target proteins or other Ub through a bond between the conserved C-terminus of Ub and the ε-amino group of a lysine residue on the target protein or other Ub (Hershko and Ciechanover, 1998). A minimum of four tandemly-attached Ub are required to allow recognition of the target protein by the 26S proteasome, presumably because a tetramer of poly-Ub assumes a higher order structure that is required for recognition (Pickart, 2000). Ub has seven lysine (Lys) residues (Figure 1A) and Ub can be conjugated to several of these Lys residues. Poly-Ub chains created by conjugation through a Lys-48 linkage targets a substrate for degradation by the 26S proteasome. In contrast, poly-Ub formed by Lys-63 conjugation does not lead to proteasome-mediated degradation, but instead is associated with the regulation of endocytosis or changes in target protein function (Schnell and Hicke, 2003). Similarly, conjugation of a single Ub (mono-ubiquitination) or less than four Ub can affect protein activity, including aspects of transcriptional regulation, protein trafficking, and endocytosis (Schnell and Hicke, 2003). The functions of poly-Ub chains created with Lys-11 and Lys-29 linkages have not been determined (Aguilar and Wendland, 2003).
Ubiquitin-mediated proteolysis is the most important pathway for the degradation of nuclear and cytosolic proteins. Inactivation of the Ub proteolytic pathway inhibits the degradation of the majority of cellular proteins, regardless of whether the proteins have short or long half-lives (Rock et al., 1994). Given the central importance of ubiquitin-mediated protein degradation in a range of cellular processes, it is not surprising that Ub-mediated pathways are important for multiple aspects of C. elegans development and cellular physiology.
The 26S proteasome is a conserved chambered protease complex that is present in both the cytoplasm and the nucleus (Wojcik and DeMartino, 2003). It consists of a 20S proteasome, a central core containing proteolytic subunits, and two 19S regulatory complexes that bind to ubiquitinated substrates, cleave off ubiquitin, and then unfold and translocate the substrate into the 20S core (Pickart and Cohen, 2004). C. elegans has 14 conserved subunits that comprise the 20S core, as well as 18 conserved 19S components (Davy et al., 2001). RNAi depletion of proteasome components during larval stages produces larval arrest and lethality while RNAi depletion in adult hermaphrodites produces progeny that arrest at the one-cell stage with defective meiosis I, indicating the central importance of this pathway (Takahashi et al., 2002; Gonczy et al., 2000).
There are two loci for ubiquitin (Ub) in C. elegans, ubq-1 and ubq-2. ubq-1 is a polyubiquitin locus (Graham et al., 1989). The predominant splice form of ubq-1 encodes an 838 amino acid peptide that includes 11 tandem Ub sequences (Figure 1B). The polyubiquitin structure of the locus is common to other eukaryotic species (Schlesinger and Bond, 1987). The polyubiquitin protein is post-translationally cleaved into individual Ub peptides by ubiquitin C-terminal hydrolases (Johnston et al., 1999). The Ub peptides of ubq-1 are identical with the exception of repeat 6, which substitutes an isoleucine for a highly conserved threonine at position 9 of the repeat (Figure 1B). The functional significance of this altered Ub peptide is not known. In C. briggsae, this atypical Ub repeat is not present, instead the orthologous polyubiquitin locus comprises ten Ub repeats that are all identical to the predominant C. elegans Ub sequence.
The second Ub locus, ubq-2, includes an intact canonical Ub fused to the L40 ribosomal large subunit protein (Jones and Candido, 1993). This fusion gene is broadly conserved in eukaryotes (Schlesinger and Bond, 1987). The ubq-2 locus contains the only copy of the L40 ribosomal subunit in the C. elegans genome. In yeast, the hybrid protein is rapidly cleaved to form Ub and the L40 ribosomal subunit (Finley et al., 1989). The transient presence of Ub in the fusion protein promotes the incorporation of the L40 subunit into ribosomes (Finley et al., 1989). Once cleaved, the Ub is functional for covalent attachment to proteins (Ozkaynak et al., 1987).
RNAi of ubq-1 or ubq-2 produces a one-cell stage arrest during the meiotic divisions, similar to inactivation of the proteasome (Gonczy et al., 2000; Piano et al., 2000). The relative importance of ubq-1 vs ubq-2 is not known, as RNAi is expected to inactivate both genes due to their extensive homology (Tijsterman et al., 2002).
As in other eukaryotes, there is only a single ubiquitin-activating enzyme in C. elegans, UBA-1. Disruption of UBA-1 activity would be expected to completely inactivate the Ub proteolytic pathway. However, while RNAi of uba-1 produces an embryonic arrest, it is not as penetrant as RNAi for ubq-1 or particular proteasome components, perhaps because, as an enzyme, it is more resistant to effects of depletion (Maeda et al., 2001; Kamath et al., 2003; Simmer et al., 2003; Piano et al., 2000; Gonczy et al., 2000).
There are 22 proteins with homology to ubiquitin-conjugating enzymes (UBCs) in C. elegans, with an additional three ubiquitin E2 variants (UEVs) that lack the critical cysteine residue in the catalytic site (Jones et al., 2002). The C. elegans UBCs are numbered ubc-1-3, 6-9, and 12-26; with numbers 4, 5, 10, and 11 skipped. The numbering of UBCs in C. elegans does not match that of S. cerevisiae or humans, and orthologous groupings are presented in Table 1. Note that ubc-9 and ubc-12 designate conjugating enzymes for the Ub-like proteins SUMO (SMO-1) and Nedd8 (NED-8), respectively, and do not conjugate Ub (Jones and Candido, 2000; Jones et al., 2002). The functions of the E2 genes have been studied in systematic RNAi screens. Only two of the 20 E2 enzymes that are specific for Ub-conjugation are essential for embryonic viability, ubc-2/let-70 and ubc-14 (Table 1; Jones et al., 2002). This is surprising given the relatively large number of E3 genes that are associated with embryonic lethal phenotypes (see below). This suggests either that UBC-2 and UBC-14 are the only E2s that function with these essential E3s or that there is significant redundancy of E2 function. In general, very little is known about which C. elegans E2s function with particular E3s. Of the remaining Ub-specific E2 genes, the RNAi depletion of four are associated with post-embryonic phenotypes: ubc-19 RNAi produces unhealthy larvae; ubc-20 RNAi produces an impenetrant L3 or L4 larval arrest; ubc-25 RNAi produces defects in neuromuscular function; and ubc-18 RNAi produces animals that have slightly slower growth and reduced brood sizes but otherwise appear wild-type (Maeda et al., 2001; Jones et al., 2002; Schulze et al., 2003; Fay et al., 2003; Table 1). Interestingly, ubc-18 functions redundantly with lin-35 Rb to promote normal pharyngeal morphogenesis, and the simultaneous inactivation of both genes causes synthetic embryonic lethality (Fay et al., 2003). For the remaining 14 E2s, inhibition by RNAi was not associated with any apparent defects.
Table 1. Ubiquitin-conjugating enzymes in C. elegans: homologs and loss-of-function phenotypes.
| C. elegans | Peptide- conjugated | S. cerevisiae | Drosophila | Human | Phenotypes | References |
|---|---|---|---|---|---|---|
| ubc-1 | Ub | UBC2 | UbcD6 | UBE2A; UBE2B | WT (RNAi) | Jones et al., 2002; Kamath et al., 2003 |
| ubc-2/let-70 | Ub | UBC4; UBC5 | effete; Dsi/Ubc1 | UBE2D1/ UBCH5A; UBE2D2/ UBCH5B; UBE2D3/ UBCH5C | embryonic arrest at pre-comma stage (RNAi) | Jones et al., 2002 |
| ubc-3 | Ub | UBC3/CDC34 | CG7656 | CDC34; FLJ20419 | WT (RNAi) | Jones et al., 2002; Fraser et al., 2000 |
| ubc-6 | Ub | UBC6 | CG5823 | NCUBE1 | WT (RNAi) | Jones et al., 2002; Kamath et al., 2003 |
| ubc-7 | Ub | UBC3 | CG9602 | UBE2G1 | WT (RNAi) | Jones et al., 2002; Kamath et al., 2003; Gonczy et al., 2000; Maeda et al., 2001 |
| ubc-8 | Ub | UBC8 | CG2257; CG14739 | UBE2H | WT (RNAi) | Jones et al., 2002 |
| ubc-9 | SUMO | UBC9 | lesswright | UBE2I | embryonic arrest post- gastrulation before muscle movement (RNAi) | Jones et al., 2002 |
| ubc-12 | NED-8 (Nedd8) | UBC12 | CG7375 | UBE2M | embryonic arrest at the comma stage (RNAi) | Jones et al., 2002 |
| ubc-13 | Ub | UBC13 | bendless; CG3473 | UBE2N; BAA93711 | WT (RNAi) | Jones et al., 2002 |
| ubc-14 | Ub | UBC7 | courtless | UBE2G2 | embryonic arrest post- gastrulation before muscle movement (RNAi) | Jones et al., 2002 |
| ubc-15 | Ub | UBC6 | CG5823 | NCUBE1 | WT (RNAi) | Jones et al., 2002; Kamath et al., 2003 |
| ubc-16 | ? | - | CG7220 | BAA91954 | WT (RNAi) | Jones et al., 2002; Fraser et al., 2000 |
| ubc-17 | ? | - | CG6303 | BAB14320; BAB14724 | WT (RNAi) | Jones et al., 2002; Kamath et al., 2003 |
| ubc-18 | Ub | - | CG17030; UbcD10; Ubc84D | UBE2L1; UBE2L3/ UBCH7; UBE2L6 | reduced growth rate and brood size (mut) | Fay et al., 2003 |
| ubc-19 | ? | - | - | - | unhealthy larvae (RNAi) | Maeda et al., 2001 |
| ubc-20 | Ub | UBC1 | UbcD4 | HIP2 | impenetrant L3 & L4 larval arrest (RNAi) | Jones et al., 2002 |
| ubc-21 | Ub | UBC1 | UbcD4 | HIP2 | WT (RNAi) | Jones et al., 2002; Kamath et al., 2003 |
| ubc-22 | Ub | - | CG17030; UbcD10; Ubc84D | UBE2L1; UBE2L3/ UBCH7; UBE2L6 | WT (RNAi) | Jones et al., 2002; Kamath et al., 2003 |
| ubc-23 | Ub | UBC1 | UbcD4 | HIP2 | WT (RNAi) | Jones et al., 2002; Kamath et al., 2003 |
| ubc-24 | ? | - | - | - | WT (RNAi) | Jones et al., 2002; Kamath et al., 2003 |
| ubc-25 | ? | - | CG2924 | UBE2Q1; UBE2Q2 | defective postembryonic neuromuscular function (RNAi) | Schulze et al., 2003 |
| ubc-26 | Ub | UBC6 | CG5823 | NCUBE1 | - | - |
| Orthologous groupings are based on published phylogenetic analysis (Jones et al., 2002; Schulze et al., 2003), with updates of other species homolog names. ubc-26 (Y110A2AM.3), was named in this study. The peptide predicted to be conjugated (Ub or Ub-like) is derived from information of homologs in other species when not known in C. elegans. Only the more severe phenotypes are listed. Phenotypes derived from RNAi or mutant analysis are denoted (brackets). WT = wild-type phenotype. References for the phenotypes listed are given. | ||||||
There are four major classes of ubiquitin ligases: HECT-domain proteins; U-box proteins; monomeric RING finger proteins; and multisubunit complexes that contain a RING finger protein (Passmore and Barford, 2004). HECT-domain E3s are unique in that Ub is transferred to a conserved cysteine residue of the E3 in a thiolester linkage, and then the E3 transfers the Ub to the substrate (Passmore and Barford, 2004; Figure 2). This function as a covalent intermediary in the transfer of Ub is not found in other classes of E3 proteins. The RING finger motif (Really Interesting New Gene) comprises eight cysteine or histidine residues that bind two Zn2+ ions in a cross-brace structure (Fang et al., 2003). The U-box is structurally similar to the RING finger motif, but it does not bind Zn2+, instead, hydrogen bonds take the place of the Zn2+ in the structure (Fang et al., 2003). Monomeric RING finger E3s and U-box E3s bind to both the substrate and the E2 enzyme (Figure 2). In multimeric RING finger complexes, the RING finger protein binds the E2 while other proteins in the complex bind the substrate. These multimeric complexes fall into two classes: cullin-based complexes, and the APC/C (anaphase promoting complex/cyclosome), which contains the cullin-like protein APC2 (Vodermaier, 2004).
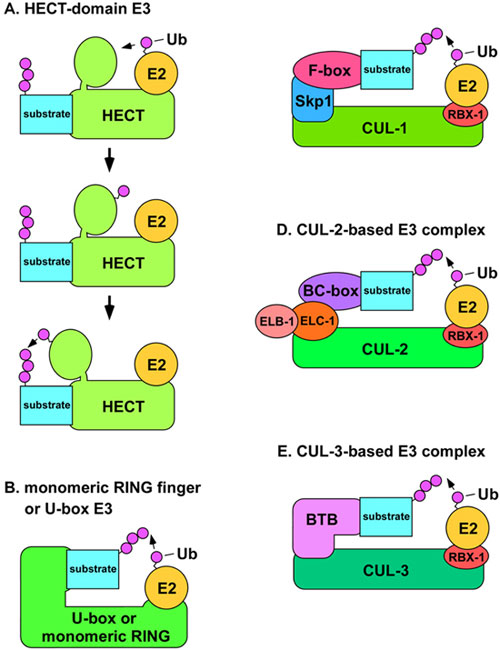
Figure 2. A) Structural model of HECT-domain E3 complex. The mechanism for the conjugation of Ub to a substrate by a HECT-domain E3 is shown. The E2 binds the N-terminal lobe of the HECT-domain E3 (top) and transfers Ub to the C-terminal lobe via a thiolester linkage (middle). The C-terminal lobe swivels on a hinge-loop and catalyzes the transfer of Ub to the substrate protein (bottom). B) Model of U-box or monomeric RING finger E3s. The U-box or RING finger domains of the E3 are directly involved in binding the E2. C) Model of SCF complexes. The N-terminus of CUL-1 binds the adaptor Skp1 (SKR proteins in C. elegans), while the C-terminus binds the RING finger protein Rbx1, which binds the E2. The substrate recognition subunit (SRS) binds to Skp1 through an F-box motif. D) Model of CUL-2-based E3 complexes. The N-terminus of CUL-2 binds to the adaptor elongin C (ELC-1), which is in complex with elongin B (ELB-1). The SRS binds to elongin C through a BC-box motif. E) Model of CUL-3-based E3 complexes. The N-terminus of CUL-3 binds directly to the SRS, which utilizes a BTB/POZ domain to bind to CUL-3.
There are nine genes in C. elegans that encode proteins with a HECT domain (Table 2). Of these genes, only two have been studied in detail: oxi-1 and wwp-1. oxi-1 was cloned as a gene whose expression increases under oxidative stress (growth in high oxygen concentrations; Yanase and Ishi, 1999). There is no observed RNAi phenotype of oxi-1; however, the RNAi analysis was not performed under high oxygen conditions when oxi-1 would be assumed to be active (Table 2). wwp-1 encodes a conserved protein with HECT and WW domains, and its RNAi depletion causes embryonic lethality with defective morphogenesis after the comma stage (Huang et al., 2000). Large-scale RNAi screens revealed that RNAi depletion of the HECT-domain gene D2085.4 produces sterility in the P0 (Maeda et al., 2001). The remaining six HECT-domain genes were not associated with reproducible RNAi phenotypes (Table 2).
Table 2. HECT-domain encoding genes in C. elegans.
| Cosmid designation | Gene name | Phenotype | References |
|---|---|---|---|
| C34D4.14 | - | WT (RNAi) | Kamath et al., 2003 |
| D2085.4 | - | P0 sterile (RNAi) | Maeda et al., 2001 |
| F36A2.13 | - | WT (RNAi) | Jones et al., 2002 |
| F45H7.6 | - | WT (RNAi) | Kamath et al., 2003 |
| Y39A1C.2 | oxi-1 | WT (RNAi) | Kamath et al., 2003; Gonczy et al., 2000 |
| Y48G8AL.1 | - | WT (3/4 trials in rrf-3 background); sterile (1/4 trials in rrf-3; RNAi) | Simmer et al., 2003 |
| Y65B4BR.4 | wwp-1 | late stage embryonic arrest with defects in morphogenesis | Huang et al., 2000 |
| Y67D8C.5 | - | embryonic lethal (10%); WT (90%; RNAi) | Maeda et al., 2001 |
| Y92H12A.2 | - | WT (RNAi) | Maeda et al., 2001 |
| Only the more severe phenotypes are listed. Phenotypes derived from RNAi or mutant analysis are denoted (brackets). WT = wild-type phenotype. References for the phenotype listed are given. | |||
There are four genes in C. elegans that encode proteins with a U-box domain (Table 3). The C. elegans U-box protein CHN-1 is the homolog of mammalian CHIP. CHIP binds to the chaperones Hsp70 and Hsp90 and functions as an E3 to degrade misfolded proteins (Hatakeyama and Nakayama, 2003). C. elegans CHN-1 also binds to the Hsp70 homolog HSP-1, suggesting a similar function (Hoppe et al., 2004). Animals homozygous for a null allele of chn-1 have slightly lower brood sizes at 20°C, but otherwise appear normal. Consistent with a cellular role with heat shock proteins, chn-1 homozygotes are sensitive to heat-stress, exhibiting larval arrest and lethality at higher temperatures (Hoppe et al., 2004; Table 3).
Table 3. U-box-domain encoding genes in C. elegans.
| Cosmid designation | Gene name | Phenotype | References |
|---|---|---|---|
| F59E10.2 | cyp-4/mog-6 | masculinization of the germ line; embryonic arrest (mut) | Graham et al., 1993 |
| T05H10.5 | ufd-2 | WT (RNAi) | Kamath et al., 2003; Piano et al., 2000 |
| T09B4.10 | chn-1 | slightly lower brood size at 20°; larval arrest at higher temperatures (mut) | Hoppe et al., 2004 |
| T10F2.4 | - | embryonic lethal (RNAi) | Kamath et al., 2003; Simmer et al., 2003; Gonczy et al., 2000 |
| Only the more severe phenotypes are listed. Phenotypes derived from RNAi or mutant analysis are denoted (brackets). WT = wild-type phenotype. References for the phenotype listed are given. | |||
C. elegans CHN-1 physically interacts with a second U-box protein, UFD-2 (Hoppe et al., 2004). UFD-2 is the ortholog of budding yeast Ufd2, which functions as an E4 (Koegl et al., 1999). As described above, an E4 enzyme catalyzes the elongation of ubiquitin chains on proteins that already have one or a few conjugated Ub (Koegl et al., 1999). Hoppe et al., found that both C. elegans CHN-1 and UFD-2 can function independently of each other as E3s for the addition of one to three Ub to UNC-45, a myosin-directed chaperone (Hoppe et al., 2004). However, more extensive poly-ubiquitination of UNC-45 in vitro required both CHN-1 and UFD-2, suggesting a novel mechanism in which a combination of E3s can produce E4 activity (Hoppe et al., 2004). The CHN-1-UFD-2 complex was able to function in vitro with UBC-2/LET-70, suggesting that UBC-2/LET-70 is the in vivo E2 (Hoppe et al., 2004).
The third U-box gene in C. elegans is cyp-4/mog-6, which is a homolog of human cyclophilin-60 (hCyp60/CYC4). In humans, hCyp60 has both peptidyl-prolyl cis/trans isomerase activity associated with its C-terminus and E3 activity associated with the U-box in its N-terminus (Hatakeyama and Nakayama, 2003). C. elegans CYP-4 also has both domains and exhibits protein-folding activity indicative of a functional prolyl isomerase (Page et al., 1996). Loss of cyp-4/mog-6 results in a failure of the hermaphrodite germ line to switch from producing sperm to producing oocytes, so that only sperm are produced (Graham et al., 1993). The prolyl isomerase domain of CYP-4/MOG-6 is not required for the sperm/oocyte switch, while the N-terminus, containing the U-box, is required (Belfiore et al., 2004). CYP-4/MOG-6 is also required for embryogenesis (Graham et al., 1993).
The final U-box gene is T10F2.4, which is homologous to yeast and human PRP19. Budding yeast Prp19 functions in spliceosome assembly, and human PRP19 has been shown to possess E3 activity (Blanton et al., 1992; Hatakeyama and Nakayama, 2003). Large-scale RNAi screens revealed that T10F2.4 is required for embryonic viability (Table 3).
There are 152 RING finger proteins in the C. elegans genome (Table 4). While a majority of RING finger proteins tested in vitro exhibit E3 activity, it is unclear if all RING finger proteins function as E3s in vivo (Fang et al., 2003). There are two classes of RING finger motifs, H2 and HC, based on the placement of His or Cys residues in positions 4 and 5 of the motif (Fang et al., 2003). The three RING finger proteins that are known to be integral components of multisubunit complexes (RBX-1, RBX-2, and APC-11) are of the H2 class and are very small proteins of 110-135 amino acids. A recent survey of RING finger proteins in C. elegans found more to be of the HC class (90 genes) than the H2 class (13 genes; Moore and Boyd, 2004). The majority of RING finger genes of either class encode proteins that are much larger than the multisubunit E3 RING-H2 proteins, as would be expected for proteins that function as monomeric E3s that bind to both the E2 and the substrate.
Table 4. RING finger encoding genes in C. elegans.
| Cosmid designation | Gene name | Phenotype | References |
|---|---|---|---|
| B0281.3 | - | WT (RNAi) | Kamath et al., 2003 |
| B0281.8 | - | WT (RNAi) | Kamath et al., 2003 |
| B0393.6 | - | Emb (RNAi) | Kamath et al., 2003; Simmer et al., 2003 |
| B0416.4 | - | WT (RNAi) | Kamath et al., 2003 |
| B0432.13 | - | WT (RNAi) | Kamath et al., 2003 |
| C01B7.6 | rpm-1, rpm-3, sam-1, sad-3, syd-3 | defective synapse formation and morphology (mut) | Schaefer et al., 2000; Zhen et al., 2000 |
| C01G6.4 | - | WT (RNAi) | Kamath et al., 2003; Moore and Boyd, 2004 |
| C02B8.6 | - | WT (RNAi) | Maeda et al., 2001 |
| C06A5.8 | - | WT (RNAi) | Jones et al., 2002; Maeda et al., 2001 |
| C06A5.9 | rnf-1, tag-54 | WT (RNAi) | Jones et al., 2002 |
| C09E7.5 | - | WT (RNAi) | Gonczy et al., 2000 |
| C09E7.8 | - | WT (RNAi) | Kamath et al., 2003; Gonczy et al., 2000 |
| C09E7.9 | - | - | - |
| C11H1.3 | - | Adl; Mlt; Unc; Dpy; Gro; Pvl (RNAi) | Kamath et al., 2003; Simmer et al., 2003 |
| C12C8.3 | lin-41 | heterochronic defect in which hypodermal cells adopt the adult fate at the L3/L4 molt (mut) | Slack et al., 2000 |
| C15F1.5 | - | WT (RNAi) | Kamath et al., 2003; Moore and Boyd, 2004 |
| C16C10.5 | - | WT (RNAi) | Kamath et al., 2003; Gonczy et al., 2000 |
| C16C10.7 | rnf-5 | disorganized body wall muscle dense bodies but normal movement (mut) | Broday et al., 2004 |
| C17E4.3 | - | Emb (RNAi) | Piano et al., 2000 |
| C17H11.6 | - | WT (RNAi) | Kamath et al., 2003 |
| C18B12.4 | - | WT (RNAi) | Kamath et al., 2003; Moore and Boyd, 2004 |
| C18H9.7 | rpy-1, rap-1 | WT (RNAi) | Kamath et al., 2003 |
| C26B9.6 | - | WT (RNAi) | Kamath et al., 2003 |
| C28G1.5 | - | - | - |
| C28G1.6 | - | - | - |
| C30F2.2 | - | WT (RNAi) | Kamath et al., 2003 |
| C32D5.10 | - | Lva (RNAi) | Moore and Boyd, 2004 |
| C32D5.11 | - | WT (RNAi) | Kamath et al., 2003; Maeda et al., 2001 |
| C32E8.1 | - | WT (RNAi) | Jones et al., 2002 |
| C34E10.4 | wrs-2 | Gro (RNAi) | Kamath et al., 2003; Simmer et al., 2003; Gonczy et al., 2000 |
| C34F11.1 | - | WT (RNAi) | Kamath et al., 2003; Maeda et al., 2001 |
| C36A4.8 | brc-1 | Him phenotype; elevated levels of germ cell death; germ cell chromosome fragmentation upon irradiation (RNAi) | Boulton et al., 2004 |
| C36B1.9 | - | - | - |
| C39F7.2 | - | WT (RNAi) | Kamath et al., 2003 |
| C45G7.4 | - | WT (RNAi) | Kamath et al., 2003 |
| C49H3.5 | ntl-4 | Emb; Gro; Slu (RNAi) | Maeda et al., 2001; Simmer et al., 2003 |
| C52E12.1 | - | - | - |
| C53A5.6 | - | Sck Ste; Lva Lvl Ste (RNAi) | Kamath et al., 2003; Simmer et al., 2003 |
| C53D5.2 | - | WT (RNAi) | Jones et al., 2002 |
| C55A6.1 | - | WT (RNAi) | Kamath et al., 2003 |
| C56A3.4 | - | Gro (RNAi) | Kamath et al., 2003; Moore and Boyd, 2004 |
| D2089.2 | - | Emb; Lvl; Pch; Slu; Gro; Unc (RNAi) | Kamath et al., 2003; Simmer et al., 2003 |
| EEED8.16 | - | - | - |
| F08B12.2 | prx-12 | Clr; L1 stage larval arrest (RNAi) | Kamath et al., 2003; Petriv et al., 2002; Thieringer et al., 2003 |
| F08G12.5 | - | WT (RNAi) | Kamath et al., 2003; Moore and Boyd, 2004 |
| F10D7.5 | - | Emb; Sck; Ste (RNAi) | Maeda et al., 2001 |
| F10G7.10 | - | WT (RNAi) | Kamath et al., 2003 |
| F11A10.3 | - | WT (RNAi) | Moore and Boyd, 2004 |
| F16A11.1 | - | WT (RNAi) | Jones et al., 2002; Maeda et al., 2001 |
| F19G12.1 | - | WT (RNAi) | Kamath et al., 2003 |
| F23B2.10 | - | - | - |
| F26E4.11 | - | Emb (RNAi) | Simmer et al., 2003 |
| F26F4.7 | nhl-2 | Ste; Stp (RNAi) | Maeda et al., 2001 |
| F26G5.9 | tam-1 | expression of genes in non-complex transgenic arrays is reduced (mut) | Hsieh et al., 1999 |
| F32A6.3 | - | Emb (RNAi) | Piano et al., 2002 |
| F35G12.9 | apc-11 | one-cell stage arrest during meiosis I; mitotic delays in escapers (RNAi) | Gonczy et al., 2000; Davis et al., 2002; Moore and Boyd, 2004 |
| F36F2.3 | - | Emb; Led (RNAi) | Jones et al., 2002; Simmer et al., 2003 |
| F40G9.12 | - | WT (RNAi) | Kamath et al., 2003; Gonczy et al., 2000; Moore and Boyd, 2004 |
| F40G9.14 | - | WT (RNAi) | Kamath et al., 2003; Gonczy et al., 2000 |
| F42C5.4 | - | WT (RNAi) | Kamath et al., 2003 |
| F42G2.5 | - | WT (RNAi) | Kamath et al., 2003 |
| F43C11.7 | - | - | - |
| F43C11.8 | - | - | - |
| F43G6.8 | - | WT (RNAi) | Kamath et al., 2003; Maeda et al., 2001 |
| F44D12.10 | - | WT (RNAi) | Kamath et al., 2003 |
| F45G2.6 | trf-1 | WT (RNAi) | Gonczy et al., 2000 |
| F46F2.1 | - | WT (RNAi) | Kamath et al., 2003 |
| F47G9.4 | - | WT (RNAi) | Kamath et al., 2003; Maeda et al., 2001 |
| F53F8.3 | - | WT (RNAi) | Kamath et al., 2003 |
| F53G2.7 | - | Emb; Ste (RNAi) | Maeda et al., 2001 |
| F54B11.5 | - | WT (RNAi) | Kamath et al., 2003 |
| F54G8.4 | nhl-1 | WT (RNAi) | Kamath et al., 2003; Gonczy et al., 2000 |
| F55A3.1 | - | WT (RNAi) | Kamath et al., 2003 |
| F55A11.3 | - | none | Moore and Boyd, 2004 |
| F55A11.7 | - | WT (RNAi) | Kamath et al., 2003 |
| F56D2.2 | - | WT (RNAi) | Kamath et al., 2003; Gonczy et al., 2000 |
| F58B6.3 | par-2 | defective embryonic anterior-posterior polarity (mut; RNAi) |
For review: |
| F58E6.1 | - | WT (RNAi) | Kamath et al., 2003; Maeda et al., 2001 |
| H05L14.2 | - | WT (RNAi) | Jones et al., 2002 |
| H10E21.5 | - | WT (RNAi) | Kamath et al., 2003; Gonczy et al., 2000 |
| K01G5.1 | - | Emb; Lva (RNAi) | Simmer et al., 2003; Gonczy et al., 2000; Moore and Boyd, 2004 |
| K02B12.8 | zhp-3 | Him (RNAi) | Jones et al., 2002; Piano et al., 2002 |
| K04C2.4 | brd-1 | Him phenotype; elevated levels of germ cell death; germ cell chromosome fragmentation upon irradiation (RNAi) | Boulton et al., 2004 |
| K09F6.7 | - | WT (RNAi) | Kamath et al., 2003 |
| K11D12.9 | - | - | - |
| K12B6.8 | - | WT (RNAi) | Kamath et al., 2003 |
| M02A10.3 | sli-1 | suppress hypomorphic alleles of let-23; low penetrance head morphology defect (mut) | Jongeward et al., 1995 |
| M88.3 | - | WT (RNAi) | Kamath et al., 2003; Gonczy et al., 2000 |
| M110.3 | - | WT (RNAi) | Kamath et al., 2003 |
| M142.6 | - | Clr; Gro (RNAi) | Kamath et al., 2003; Moore and Boyd, 2004 |
| R02E12.4 | - | WT (RNAi) | Kamath et al., 2003 |
| R05D3.4 | rfp-1 | Gro; Lva; Pvl; Rup; Stp; Unc; Egl (RNAi) | Kamath et al., 2003; Piano et al., 2002; Simmer et al., 2003; Crowe and Candido, 2004 |
| R06F6.2 | - | Bmd; Lvl; Mlt; Emb; Gro; Sma (RNAi) | Kamath et al., 2003; Simmer et al., 2003; |
| R10A10.2 | rbx-2 | WT (RNAi) | Jones et al., 2002; Maeda et al., 2001; Piano et al., 2002; Moore and Boyd, 2004 |
| T01C3.3 | - | WT (RNAi) | Kamath et al., 2003; Piano et al., 2002; Moore and Boyd, 2004 |
| T01G5.7 | - | WT (RNAi) | Kamath et al., 2003 |
| T02C1.1 | - | WT (RNAi) | Kamath et al., 2003; Gonczy et al., 2000 |
| T02C1.2 | - | - | - |
| T05A12.4 | - | WT (RNAi) | Kamath et al., 2003; Maeda et al., 2001 |
| T08D2.4 | - | WT (RNAi) | Kamath et al., 2003; Moore and Boyd, 2004 |
| T13A10.2 | - | - | - |
| T13H2.5 | - | - | - |
| T20F5.6 | - | WT (RNAi) | Jones et al., 2002; Piano et al., 2002 |
| T20F5.7 | - | WT (RNAi) | Jones et al., 2002 |
| T22B2.1 | - | WT (RNAi) | Kamath et al., 2003 |
| T23F6.3 | - | WT (RNAi) | Kamath et al., 2003 |
| T24D1.2 | - | WT (RNAi) | Jones et al., 2002; Piano et al., 2002; Moore and Boyd, 2004 |
| T24D1.3 | - | Emb (RNAi) | Piano et al., 2002 |
| T24D1.5 | - | - | - |
| T26C12.3 | - | WT (RNAi) | Kamath et al., 2003; Maeda et al., 2001 |
| W02A11.3 | - | WT (RNAi) | Jones et al., 2002; Moore and Boyd, 2004 |
| W04H10.3 | nhl-3 | WT (RNAi) | Kamath et al., 2003 |
| W09G3.6 | - | WT (RNAi) | Jones et al., 2002 |
| Y4C6A.3 | - | WT (RNAi) | Kamath et al., 2003 |
| Y6D1A.2 | - | WT (RNAi) | Maeda et al., 2001 |
| Y7A9C.1 | - | WT (RNAi) | Kamath et al., 2003 |
| Y38C1AA.6 | - | - | - |
| Y38F1A.2 | - | - | - |
| Y38H8A.2 | - | - | - |
| Y45F10B.8 | - | WT (RNAi) | Kamath et al., 2003 |
| Y45F10B.9 | - | WT (RNAi) | Kamath et al., 2003 |
| Y45G12B.2 | - | - | - |
| Y47D3A.22 | - | - | - |
| Y47D3B.11 | - | - | - |
| Y47G6A.14 | - | WT (RNAi) | Jones et al., 2002 |
| Y51F10.2 | - | - | - |
| Y52E8A.2 | - | - | - |
| Y53G8AM.4 | - | - | - |
| Y54E10A.11 | - | - | - |
| Y54E10BR.3 | - | WT (RNAi) | Jones et al., 2002 |
| Y55F3AM.6 | - | WT (RNAi) | Kamath et al., 2003 |
| Y57A10B.1 | - | WT (RNAi) | Kamath et al., 2003 |
| Y67D8B.1 | - | - | - |
| Y71F9AL.10 | - | - | - |
| Y73C8C.7 | - | WT (RNAi) | Kamath et al., 2003 |
| Y73C8C.8 | - | WT (RNAi) | Kamath et al., 2003 |
| Y75B8A.10 | - | WT (RNAi) | Kamath et al., 2003; Gonczy et al., 2000 |
| Y105C5B.11 | - | - | - |
| Y105E8A.14 | - | WT (RNAi) | Jones et al., 2002 |
| Y119C1B.5 | - | - | - |
| ZC13.1 | - | WT (RNAi) | Kamath et al., 2003 |
| ZK287.5 | rbx-1 | one cell stage embryonic arrest (RNAi) | Moore and Boyd, 2004; Sasagawa et al., 2003 |
| ZK637.14 | - | WT (RNAi) | Kamath et al., 2003; Gonczy et al., 2000 |
| ZK809.7 | prx-2 | Gro (RNAi) | Kamath et al., 2003; Simmer et al., 2003; |
| ZK993.2 | - | - | - |
| ZK1240.1 | - | Lvl; Sck; Gro; Unc (RNAi) | Kamath et al., 2003; Maeda et al., 2001; Simmer et al., 2003 |
| ZK1240.2 | - | WT (RNAi) | Kamath et al., 2003 |
| ZK1240.3 | - | WT (RNAi) | Kamath et al., 2003 |
| ZK1240.6 | - | WT (RNAi) | Kamath et al., 2003; Moore and Boyd, 2004 |
| ZK1240.8 | - | - | - |
| ZK1240.9 | - | - | - |
| ZK1320.6 | arc-1, arl-4 | WT (RNAi) | Maeda et al., 2001; Moore and Boyd, 2004 |
| Only the more severe phenotypes are listed. Phenotypes derived from RNAi or mutant analysis are denoted (brackets). Phenotype abbreviations are defined in WormBase. References for the phenotype listed are given. | |||
Twenty one of the RING finger proteins in C. elegans that do not function as components of known multisubunit E3 complexes have been the focus of genetic studies. (see named genes in Table 4). Most of these have not been studied for possible E3 activity. Five of the potentially monomeric RING finger proteins have been implicated as E3s, and are discussed below.
RNF-5 is the ortholog of the mammalian E3 RNF5 (Didier et al., 2003). RNAi depletion of rnf-5 causes a disorganization of body wall muscle dense bodies (Broday et al., 2004). RNF-5 binds and negatively regulates the protein level of UNC-95, a LIM-domain protein that is required for the integrity of dense bodies (Didier et al., 2003; Broday et al., 2004). Mutations of the RING finger domain of RNF-5 severely reduce its ability to lower UNC-95 protein levels upon overexpression (Broday et al., 2004). These results suggest that RNF-5 directly targets UNC-95 for ubiquitin-mediated proteolysis.
SLI-1 is the ortholog of mammalian c-Cbl. c-Cbl functions as an E3 to ubiquitinate active receptor tyrosine kinases (RTKs) to induce their endocytosis or degradation (Shtiegman and Yarden, 2003). SLI-1 negatively regulates the LET-23 RTK, which mediates vulva differentiation (Yoon et al., 1995). A sli-1 mutant that lacks the RING finger domain has a significantly reduced ability to inhibit LET-23 activity, suggesting that SLI-1 functions as an E3 to regulate LET-23 (Yoon et al., 2000).
RFP-1 was identified as a binding partner for the E2 UBC-1 in a yeast two-hybrid screen (Crowe and Candido, 2004). RNAi depletion of rfp-1 produces an L1-stage larval arrest, while escapers exhibit vulval and egg-laying defects (Crowe and Candido, 2004). Interestingly, RNAi depletion of ubc-1, the predicted E2, does not produce any phenotypes, suggesting that RFP-1 can function with additional E2s (Crowe and Candido, 2004).
BRC-1 and BRD-1 are RING finger proteins that are orthologs of the mammalian breast cancer susceptibility gene BRCA1 and the BRCA1-associating protein BRD-1, respectively (Boulton et al., 2004). In mammals, a complex of BRCA1 and BRD1 exhibits E3 activity in vitro, and mutations in the RING finger of BRCA1 that abolish E3 activity are associated with breast cancer (Ohta and Fukuda, 2004). In C. elegans, BRC-1 also binds BRD-1 (Boulton et al., 2004). RNAi depletion of either gene produces a high incidence of males (Him) phenotype, elevated levels of p53-dependent germ cell death, and chromosome fragmentation after irradiation, suggesting a role in DNA repair (Boulton et al., 2004). Intriguingly, mammalian BRCA1 and BRD1 have been implicated in the ubiquitination of p53 (Dong et al., 2003), suggesting that the p53-dependent germ cell death observed upon BRC-1 or BRD-1 RNAi results from elevated p53 levels. In concordance with this, loss of p53 (cep-1) suppressed the brc-1 and brd-1 germ cell deaths (Boulton et al., 2004).
RPM-1 encodes a protein with RING finger and guanine nucleotide exchange domains. rpm-1 mutants have varied defects in neuron branching, and synapse organization and structure, indicating a critical role in presynaptic development (Schaefer et al., 2000; Zhen et al., 2000). RPM-1 controls presynaptic development by negatively regulating DLK-1, which functions as the initial MAP kinase (MAPK) in a MAPK cascade (Nakata et al., 2005). Co-expression of RPM-1 and DLK-1 in mammalian cells promotes the ubiquitination of DLK-1, and DLK-1 is co-immunoprecipitated by the C-terminus of RPM-1, suggesting that RPM-1 directly binds and mediates DLK-1 ubiquitination (Nakata et al., 2005). These results identified the first ubiquitinated substrate of an RPM-1 family member, which also control synapse formation in Drosophila and mammals (Chang and Balice-Gordon, 2000; Burgess et al., 2004). RPM-1 has also been shown to physically associate with an SCF E3 complex (Liao et al., 2004), and this will be discussed in the CUL-1 section of the review.
Cullins are a conserved family of E3 components that were first identified in C. elegans and budding yeast (Kipreos et al., 1996; Mathias et al., 1996). There are six cullins in C. elegans. The crystal structure of a human CUL1-based SCF complex reveals that CUL1 forms a rigid scaffold in which the C-terminus binds the RING-H2 finger protein Rbx1/Roc1, and the N-terminus binds to the adaptor Skp1 (Zheng et al., 2002; Figure 2). Skp1 binds the substrate-recognition subunit (SRS), through an F-box motif of the SRS. The SRS binds substrates and positions them for ubiquitination. The E2 binds the complex through interaction with Rbx1 (Figure 2). Other cullins are also predicted to function as scaffolds with similar tertiary structure (Wu et al., 2003). All cullin-based E3s components interact with multiple SRSs, each of which form distinct E3 complexes that direct the ubiquitination of distinct sets of substrates (Guardavaccaro and Pagano, 2004). Based on the large number of potential SRS genes in metazoan genomes, cullin-based E3s may comprise the most abundant class of E3 in metazoa.
Each class of cullin-based E3 complexes includes a small RING finger protein, either Rbx1/Roc1 or Rbx2/Roc2 (Ohta et al., 1999). C. elegans RBX-1 and RBX-2 share 36% identity. Partial inactivation of the C. elegans rbx-1 gene produces phenotypes that resemble a mixture of different cullin loss-of-function phenotypes (Sasagawa et al., 2003; Moore and Boyd, 2004; data not shown). Complete inactivation of rbx-1 causes a one-cell stage arrest that is more severe than any individual cullin loss-of-function phenotype, and may reflect the simultaneous loss of multiple cullin complexes (Sasagawa et al., 2003; Moore and Boyd, 2004). In contrast, inactivation of rbx-2 by RNAi does not cause any apparent defects (Moore and Boyd, 2004), suggesting a more limited role.
Cullin-based complexes are themselves regulated by the covalent addition of a ubiquitin-like protein, Nedd8, onto the cullin. Nedd8 promotes cullin activity, at least in part, by blocking the binding of the cullin inhibitor CAND1 (Pan et al., 2004). NED-8 is removed from cullins by the COP9 Signalosome complex (CSN; Cope and Deshaies, 2003). In C. elegans, inactivation of either the Nedd8 homolog NED-8 or CSN components produce embryonic phenotypes similar to cul-3 RNAi, suggesting that both neddylation and deneddylation are required for CUL-3 activity (Pintard et al., 2003). In contrast, the early embryonic phenotypes observed in cul-2 mutants are not seen upon inactivation of NED-8 or CSN, suggesting that NED-8 modification is not essential for CUL-2 function (Pintard et al., 2003). The functional requirement for NED-8 conjugation of other cullins has not been studied.
CUL-1-based E3 complexes are known as SCF complexes to reflect the components: Skp1 (the adaptor); CUL1/Cdc53; and an F-box protein (the substrate recognition subunit; Figure 2). Distinct SCF complexes are formed by the combination of core components (Skp1, CUL1, and Rbx1) with different F-box proteins. C. elegans has an extremely large number of F-box proteins relative to other metazoa: at least 326 in C. elegans compared to 13 in budding yeast and 68 in humans (Kipreos and Pagano, 2000; Jin et al., 2004). This larger number of F-box protein genes is matched by a larger number of Skp1-related genes in C. elegans (21 SKR genes) relative to only a single Skp1 gene in budding yeast and humans (Nayak et al., 2002; Yamanaka et al., 2002). Seven of the SKRs were found to interact with CUL-1: SKR-1, -2, -3, -7, -8, -9, and -10 (Nayak et al., 2002; Yamanaka et al., 2002). The observation of multiple SKRs that can bind CUL-1, along with the large number of F-box proteins, suggests that C. elegans will contain many distinct SCF complexes. The cellular functions associated with known SCF complexes are discussed below.
SCFLIN-23: LIN-23 is an F-box protein with WD-repeats. lin-23 mutants have hyperplasia in all somatic lineages caused by a failure of dividing blast cells to cease cell division at the appropriate time (Kipreos et al., 2000). This cell cycle exit defect is also the primary phenotype associated with cul-1 mutants and with skr-1 and skr-2 RNAi, suggesting that the closely related SKR-1 and SKR-2 proteins function as adaptors in the SCFLIN-23 complex (Kipreos et al., 1996; Nayak et al., 2002). LIN-23 has a separate function to promote the proper outgrowth of axons (Mehta et al., 2004).
SCFSEL-10: SEL-10 is an F-box protein with WD-repeats that negatively regulates signaling by the Notch family member LIN-12 (Hubbard et al., 1997). SEL-10 was shown to physically bind the LIN-12 receptor (a Notch family member), suggesting that it directly mediates LIN-12 ubiquitination (Hubbard et al., 1997). This pathway was subsequently shown to be conserved in mammals, with the SEL-10 ortholog responsible for the ubiquitin-mediated degradation of Notch (Wu et al., 2001; Oberg et al., 2001). SEL-10 also binds and negatively regulates the presenilin SEL-12 (Wu et al., 1998). Studies with human cells have subsequently shown that the degradation of presenilin by SCFSEL-10 is conserved (Li et al., 2002). Finally, SEL-10 binds and facilitates the degradation of FEM-1 and FEM-3, which function in the sex determination pathway to promote male development (Jager et al., 2004).
SCFFSN-1: FSN-1 is an F-box protein with a SPRY domain that is required for regulating neuromuscular junction (synapse) formation in different classes of neurons (Liao et al., 2004). Co-immunoprecipitation (co-IP) analysis indicates that FSN-1 physically associates with CUL-1 and SKR-1, suggesting that it functions as the SRS for an SCF complex (Liao et al., 2004). FSN-1 binds and negatively regulates the level of SCD-1/ALK (Liao et al., 2004). An scd-1 mutant suppresses the fsn-1 synapse defect, suggesting that SCD-1 is the critical substrate of the SCFFSN-1 complex for regulating synapse formation (Liao et al., 2004).
FSN-1 has been shown by co-IP analysis to physically associate with the large RING finger protein RPM-1, and it has been proposed that RPM-1 functions as the RING finger component of the SCFFSN-1 complex (Liao et al., 2004). However, there is evidence that suggests that RPM-1 does not function analogously to RBX-1 in an SCFFSN-1 complex. In particular, rpm-1 null mutants are not functionally equivalent to fsn-1 null mutants, and RPM-1 requires the presence of FSN-1 to associate with SCF components, which would not be expected for a component that binds directly to CUL-1 (Liao et al., 2004).
CUL-2-based E3 complexes have a structure very similar to that of SCF complexes (Wu et al., 2003; Figure 2). CUL-2 complexes employ the Skp1-related protein elongin C as an adaptor in combination with elongin B, which contains a ubiquitin-like domain (Kim and Kaelin, 2003). Substrate recognition subunits bind elongin C through a BC-box/VHL-box motif (Kamura et al., 2004). The C. elegans cul-2 gene is the only metazoan cul-2 ortholog whose functions have been analyzed genetically. cul-2 mutants have a large number of phenotypes reflecting diverse cellular functions. 1) CUL-2 is required for the G1-to-S phase transition in germ cells, which is caused, at least in part, by a failure to negatively regulate the levels of the CDK-inhibitor CKI-1 (Feng et al., 1999). 2) CUL-2 is required for the meiosos II metaphase to anaphase transition and meiosis II exit. The failure/delay of the metaphase II to anaphase II transition is correlated with a failure to degrade the cell cycle regulator cyclin B1, while the delay in meiosis exit is correlated with a failure to degrade cyclin B3 (Liu et al., 2004; Sonneville and Gonczy, 2004). 3) CUL-2 is required for proper anterior-posterior (A-P) polarity. In cul-2 mutants, A-P polarity is often reversed due to the perduring meiotic spindle acting as a catalyst for the ectopic placement of the PAR-2 polarity protein onto the anterior cortex (Liu et al., 2004; Sonneville and Gonczy, 2004). Additionally, CUL-2 restricts the localization of PAR-2 in regions distant from microtubule-organizing centers (Liu et al., 2004; Sonneville and Gonczy, 2004). 4) CUL-2 is required for defects in mitotic chromosome condensation and mitotic progression (Feng et al., 1999). 5) CUL-2 is required to prevent cytoplasmic extensions/blebbing in the early embryo (Feng et al., 1999). 6) A CUL-2 complex containing the SRS ZIF-1 is required for the degradation of five CCCH Zn finger polarity proteins (PIE-1, POS-1, MEX-1, MEX-5, and MEX-6) in non-germ cell embryonic lineages (DeRenzo et al., 2003).
In mammals, CUL-2 functions in a complex with the von Hippel-Lindau tumor suppressor protein (VHL) as the SRS to target the degradation of hypoxia inducible factor-1α (HIF-1α; Kim and Kaelin, 2003). In C. elegans, VHL-1 also promotes HIF-1 degradation, so it is likely that C. elegans VHL-1 functions as a component of a CUL-2 complex (Epstein et al., 2001).
CUL-3 has a slightly different structure from that of CUL-1 and CUL-2-based complexes in that a single BTB/POZ-domain protein functions as both the substrate recognition subunit and adaptor, i.e., the BTB protein binds CUL-3 and the substrate (van den Heuvel, 2004; Figure 2). C. elegans contains over 100 BTB-domain proteins, indicating the possibility for multiple CUL-3-based complexes (Furukawa et al., 2003; Xu et al., 2003). C. elegans was the first organism in which a functional CUL-3-based E3 complex and its substrate were identified. A C. elegans CUL-3 complex that contains the BTB protein MEL-26 was shown to degrade the microtubule-severing katanin MEI-1 (Pintard et al., 2003; Furukawa et al., 2003; Xu et al., 2003). MEI-1 is degraded in the one-cell embryo after meiosis (Pintard et al., 2003). Presumably, the microtubule-severing activity of MEI-1 is required during meiosis to restrict the size of the meiotic spindle, but must be destroyed to allow the larger mitotic spindle to form after meiosis. In cul-3 RNAi animals or mel-26 mutants, mitotic aster microtubules are disorganized and shorter compared to wild type, and this is associated with defects in spindle positioning and elongation, and cytokinesis (Kurz et al., 2002; Pintard et al., 2003; Dow and Mains, 1998).
The structure of CUL-4-based E3 complexes has not been fully worked out. In mammals, the CUL-4 complex includes the DDB1 protein, which appears to be capable of functioning either as an adaptor or as a substrate recognition subunit (Wertz et al., 2004; Hu et al., 2004). In C. elegans, CUL-4 has a central role in the regulation of DNA replication by restricting replication to only once per cell cycle (Zhong et al., 2003). RNAi depletion of cul-4 produces an L2-stage larval arrest in which blast cells undergo unrestrained re-replication and attain elevated DNA contents up to 100 C (Zhong et al., 2003). CUL-4 is required for the degradation of the replication licensing factor CDT-1 during S phase (Zhong et al., 2003). The degradation of CDT-1 precludes it from reloading the MCM complex onto replication origins, thereby preventing the re-initiation of DNA replication at origins during the same cell cycle (Zhong et al., 2003; Feng and Kipreos, 2003). The degradation of CDT-1 by CUL-4 was subsequently shown to be conserved in Drosophila and mammals (Higa et al., 2003).
The APC/C is a conserved multisubunit E3 complex that functions during meiosis, mitosis, and G1 phase (Yeong, 2004). C. elegans contains nine core APC/C components, as well as two accessory components, FZY-1 (also known as CDC20) and FZR-1 (CDH1; Yeong, 2004). C. elegans and budding yeast were the first organisms in which a role for the APC/C in chromosome separation during meiosis I was demonstrated (Salah and Nasmyth, 2000; Yeong, 2004). Mutations of APC/CFZY-1 components produce failures of chromosome separation during mitosis and meiosis I of both oocyte and sperm lineages (Furuta et al., 2000; Davis et al., 2002; Shakes et al., 2003; Kitagawa et al., 2002). Inactivation of APC/CFZY-1 components causes a one-cell arrest at metaphase of meiosis I that is similar to what is observed upon inactivation of proteasome components, suggesting that APC/CFZY-1 mediates this initial requirement for ubiquitin proteolysis in the embryo (Furuta et al., 2000; Davis et al., 2002; Shakes et al., 2003; Kitagawa et al., 2002; Gonczy et al., 2000).
C. elegans APC/C is required for the degradation of IFY-1, the proposed Securin that functions to release Separase, SEP-1, which is a conserved protease that separates sister chromatids at anaphase (Siomos et al., 2001; Kitagawa et al., 2002). However, the degradation of Securin cannot be the sole essential function of APC/CFZY-1 in promoting meiosis I, as experimentally-inducing a loss of chromosome cohesion in APC/C mutants (which should bypass the requirement for Securin degradation) does not rescue the meiosis I arrest (Davis et al., 2002). The APC/C-mediated release of active SEP-1 has been suggested to directly affect anterior-posterior polarity in the one-cell stage embryo by ensuring that the paternal pronucleus/centrosome complex remains in tight association with the posterior cortex of the embryo, where it promotes the cortical association of the PAR-2 polarity protein (Rappleye et al., 2002). Hypomorphic alleles of APC/C or RNAi depletion of SEP-1 are associated with a lack of embryonic polarity due to a failure of PAR-2 to localize to the posterior cortex (Rappleye et al., 2002). However, another study has suggested that the polarity defects are secondary consequences of a failure of meiosis and do not imply a direct regulation of polarity by APC/C or SEP-1 (Shakes et al., 2003).
Genetic experiments have implicated APC/CFZR-1 in the negative regulation of cyclins during G1 phase (Fay et al., 2003). In mammals, the Rb protein negatively regulates the transcription of cyclins in G1 phase (Peters, 2002). Weak alleles of fzr-1 have no overt phenotypes by themselves, but produce a synthetic hyperplasia phenotype when combined with homozygous lin-35 Rb mutant alleles. Additionally, overexpression of cyclins A and E produce more extensive hyperplasia in a fzr-1 mutant background, suggesting that APC/CFZR-1 degrades S phase and mitotic cyclins during G1 phase, as occurs in other metazoa (Fay et al., 2003). More complete inactivation of fzr-1 by RNAi reveals severe pleiotropic effects on cell proliferation and development (Fay et al., 2003). Biochemical and genetic experiments indicate that APC/C functions with the E2 UBC-2 to promote both meiosis and mitosis (Frazier et al., 2004).
The APC also has a non-cell cycle function to regulate the abundance of GLR-1 glutamate receptors in ventral cord nerve cells (Juo and Kaplan, 2004). The endocytosis of GLR-1 is induced by the covalent attachment of one or a few Ub to GLR-1 (Burbea et al., 2002). APC/C promotes GLR-1 endocytosis; however, APC/C does not directly ubiquitinate GLR-1, and the critical target of APC/C in regulating GLR-1 endocytosis is not yet known (Juo and Kaplan, 2004).
Research in the Kipreos laboratory is supported by the National Institutes of Health (R01 GM55297) and the American Cancer Society (RSG-01-251-01-DDC).
Aguilar, R.C., and Wendland, B. (2003). Ubiquitin: not just for proteasomes anymore. Curr. Opin. Cell Biol. 15, 184–190. Abstract Article
Belfiore, M., Pugnale, P., Saudan, Z., and Puoti, A. (2004). Roles of the C. elegans cyclophilin-like protein MOG-6 in MEP-1 binding and germline fates. Development 131, 2935–2945. Abstract Article
Blanton, S., Srinivasan, A., and Rymond, B.C. (1992). PRP38 encodes a yeast protein required for pre-mRNA splicing and maintenance of stable U6 small nuclear RNA levels. Mol. Cell. Biol. 12, 3939–3947. Abstract
Boulton, S.J., Martin, J.S., Polanowska, J., Hill, D.E., Gartner, A., and Vidal, M. (2004). BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr. Biol. 14, 33–39. Abstract Article
Broday, L., Kolotuev, I., Didier, C., Bhoumik, A., Podbilewicz, B., and Ronai, Z. (2004). The LIM domain protein UNC-95 is required for the assembly of muscle attachment structures and is regulated by the RING finger protein RNF-5 in C. elegans. J. Cell Biol. 165, 857–867. Abstract Article
Burbea, M., Dreier, L., Dittman, J.S., Grunwald, M.E., and Kaplan, J.M. (2002). Ubiquitin and AP180 regulate the abundance of GLR-1 glutamate receptors at postsynaptic elements in C. elegans. Neuron 35, 107–120. Abstract Article
Burgess, R.W., Peterson, K.A., Johnson, M.J., Roix, J.J., Welsh, I.C., and O'Brien, T.P. (2004). Evidence for a conserved function in synapse formation reveals Phr1 as a candidate gene for respiratory failure in newborn mice. Mol. Cell Biol. 24, 1096–1105. Abstract Article
Chang, Q., and Balice-Gordon, R.J. (2000). Highwire, rpm-1, and futsch: balancing synaptic growth and stability. Neuron 26, 287–290. Abstract Article
Cope, G.A., and Deshaies, R.J. (2003). COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114, 663–671. Abstract Article
Crowe, E., and Candido, E.P. (2004). Characterization of C. elegans RING finger protein 1, a binding partner of ubiquitin-conjugating enzyme 1. Dev. Biol. 265, 446–459. Abstract Article
Davis, E.S., Wille, L., Chestnut, B.A., Sadler, P.L., Shakes, D.C., and Golden, A. (2002). Multiple subunits of the Caenorhabditis elegans anaphase-promoting complex are required for chromosome segregation during meiosis I. Genetics 160, 805–813. Abstract
Davy, A., Bello, P., Thierry-Mieg, N., Vaglio, P., Hitti, J., Doucette-Stamm, L., Thierry-Mieg, D., Reboul, J., Boulton, S., Walhout, A.J. (2001). A protein–protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep. 2, 821–828. Abstract Article
DeRenzo, C., Reese, K.J., and Seydoux, G. (2003). Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature 424, 685–689. Abstract Article
Didier, C., Broday, L., Bhoumik, A., Israeli, S., Takahashi, S., Nakayama, K., Thomas, S.M., Turner, C.E., Henderson, S., Sabe, H., and Ronai, Z. (2003). RNF5, a RING finger protein that regulates cell motility by targeting paxillin ubiquitination and altered localization. Mol. Cell Biol. 23, 5331–5345. Abstract Article
Dong, Y., Hakimi, M.A., Chen, X., Kumaraswamy, E., Cooch, N.S., Godwin, A.K., and Shiekhattar, R. (2003). Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol. Cell 12, 1087–1099. Abstract Article
Dow, M.R., and Mains, P.E. (1998). Genetic and molecular characterization of the Caenorhabditis elegans gene, mel-26, a postmeiotic negative regulator of mei-1, a meiotic-specific spindle component. Genetics 150, 119–128. Abstract
Epstein, A.C., Gleadle, J.M., McNeill, L.A., Hewitson, K.S., O'Rourke, J., Mole, D.R., Mukherji, M., Metzen, E., Wilson, M.I., Dhanda, A. (2001). C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54. Abstract Article
Fang, S., Lorick, K.L., Jensen, J.P., and Weissman, A.M. (2003). RING finger ubiquitin protein ligases: implications for tumorigenesis, metastasis and for molecular targets in cancer. Semin. Cancer Biol. 13, 5–14. Abstract Article
Fay, D.S., Large, E., Han, M., and Darland, M. (2003). lin-35/Rb and ubc-18, an E2 ubiquitin-conjugating enzyme, function redundantly to control pharyngeal morphogenesis in C. elegans. Development 130, 3319–3330. Abstract Article
Feng, H., and Kipreos, E.T. (2003). Preventing DNA re-replication–divergent safeguards in yeast and metazoa. Cell Cycle 2, 431–434. Abstract
Feng, H., Zhong, W., Punkosdy, G., Gu, S., Zhou, L., Seabolt, E.K., and Kipreos, E.T. (1999). CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat. Cell Biol. 1, 486–492. Abstract Article
Finley, D., Bartel, B., and Varshavsky, A. (1989). The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338, 394–401. Abstract Article
Fraser, A.G., Kamath, R.S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M., and Ahringer, J. (2000). Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330. Abstract Article
Frazier, T., Shakes, D., Hota, U., and Boyd, L. (2004). Caenorhabditis elegans UBC-2 functions with the anaphase-promoting complex but also has other activities. J. Cell Sci. 117, 5427–5435. Abstract Article
Furukawa, M., He, Y.J., Borchers, C., and Xiong, Y. (2003). Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 5, 1001–1007. Abstract Article
Furuta, T., Tuck, S., Kirchner, J., Koch, B., Auty, R., Kitagawa, R., Rose, A.M., and Greenstein, D. (2000). EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol. Biol. Cell 11, 1401–1419. Abstract
Gonczy, P., Echeverri, C., Oegema, K., Coulson, A., Jones, S.J., Copley, R.R., Duperon, J., Oegema, J., Brehm, M., Cassin, E. (2000). Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408, 331–336. Abstract Article
Graham, P.L., Schedl, T., and Kimble, J. (1993). More mog genes that influence the switch from spermatogenesis to oogenesis in the hermaphrodite germ line of Caenorhabditis elegans. Dev. Genet. 14, 471–484. Abstract Article
Graham, R.W., Jones, D., and Candido, E.P. (1989). UbiA, the major polyubiquitin locus in Caenorhabditis elegans, has unusual structural features and is constitutively expressed. Mol. Cell Biol. 9, 268–277. Abstract
Guardavaccaro, D., and Pagano, M. (2004). Oncogenic aberrations of cullin-dependent ubiquitin ligases. Oncogene 23, 2037–2049. Abstract Article
Hatakeyama, S., and Nakayama, K.I. (2003). U-box proteins as a new family of ubiquitin ligases. Biochem. Biophys. Res. Commun. 302, 635–645. Abstract Article
Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. Abstract Article
Higa, L.A., Mihaylov, I.S., Banks, D.P., Zheng, J., and Zhang, H. (2003). Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5, 1008–1015. Abstract Article
Hoppe, T., Cassata, G., Barral, J.M., Springer, W., Hutagalung, A.H., Epstein, H.F., and Baumeister, R. (2004). Regulation of the myosin-directed chaperone UNC-45 by a novel E3/E4-multiubiquitylation complex in C. elegans. Cell 118, 337–349. Abstract Article
Hsieh, J., Liu, J., Kostas, S.A., Chang, C., Sternberg, P.W., and Fire, A. (1999). The RING finger/B-box factor TAM-1 and a retinoblastoma-like protein LIN-35 modulate context-dependent gene silencing in Caenorhabditis elegans. Genes Dev. 13, 2958–2970. Abstract Article
Hu, J., McCall, C.M., Ohta, T., and Xiong, Y. (2004). Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6, 1003–1009. Abstract Article
Huang, K., Johnson, K.D., Petcherski, A.G., Vandergon, T., Mosser, E.A., Copeland, N.G., Jenkins, N.A., Kimble, J., and Bresnick, E.H. (2000). A HECT domain ubiquitin ligase closely related to the mammalian protein WWP1 is essential for Caenorhabditis elegans embryogenesis. Gene 252, 137–145. Abstract Article
Hubbard, E.J., Wu, G., Kitajewski, J., and Greenwald, I. (1997). sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 11, 3182–3193. Abstract
Jager, S., Schwartz, H.T., Horvitz, H.R., and Conradt, B. (2004). The Caenorhabditis elegans F-box protein SEL-10 promotes female development and may target FEM-1 and FEM-3 for degradation by the proteasome. Proc. Natl. Acad. Sci. USA 101, 12549–12554. Abstract Article
Jin, J., Cardozo, T., Lovering, R.C., Elledge, S.J., Pagano, M., and Harper, J.W. (2004). Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18, 2573–2580. Abstract Article
Johnston, S.C., Riddle, S.M., Cohen, R.E., and Hill, C.P. (1999). Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 18, 3877–3887. Abstract Article
Jones, D., and Candido, E.P. (1993). Novel ubiquitin-like ribosomal protein fusion genes from the nematodes Caenorhabditis elegans and Caenorhabditis briggsae. J. Biol. Chem. 268, 19545–19551. Abstract
Jones, D., and Candido, E.P. (2000). The NED-8 conjugating system in Caenorhabditis elegans is required for embryogenesis and terminal differentiation of the hypodermis. Dev. Biol. 226, 152–165. Abstract Article
Jones, D., Crowe, E., Stevens, T.A., and Candido, E.P. (2002). Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol. 3, RESEARCH0002. Abstract
Jongeward, G.D., Clandinin, T.R., and Sternberg, P.W. (1995). sli-1, a negative regulator of let-23-mediated signaling in C. elegans. Genetics 139, 1553–1566. Abstract
Juo, P., and Kaplan, J.M. (2004). The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Curr. Biol. 14, 2057–2062. Abstract Article
Kamath, R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237. Abstract Article
Kamura, T., Maenaka, K., Kotoshiba, S., Matsumoto, M., Kohda, D., Conaway, R.C., Conaway, J.W., and Nakayama, K.I. (2004). VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 18, 3055–3065. Abstract Article
Kim, W., and Kaelin, W.G., Jr. (2003). The von Hippel-Lindau tumor suppressor protein: new insights into oxygen sensing and cancer. Curr. Opin. Genet. Dev. 13, 55–60. Abstract Article
Kipreos, E.T., Gohel, S.P., and Hedgecock, E.M. (2000). The C. elegans F-box/WD-repeat protein LIN-23 functions to limit cell division during development. Development 127, 5071–5082. Abstract
Kipreos, E.T., Lander, L.E., Wing, J.P., He, W.W., and Hedgecock, E.M. (1996). cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 85, 829–839. Abstract Article
Kipreos, E.T., and Pagano, M. (2000). The F-box protein family. Genome Biol. 1, REVIEWS3002. Abstract Article
Kitagawa, R., Law, E., Tang, L., and Rose, A.M. (2002). The Cdc20 homolog, FZY-1, and its interacting protein, IFY-1, are required for proper chromosome segregation in Caenorhabditis elegans. Curr. Biol. 12, 2118–2123. Abstract Article
Koegl, M., Hoppe, T., Schlenker, S., Ulrich, H.D., Mayer, T.U., and Jentsch, S. (1999). A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96, 635–644. Abstract Article
Kurz, T., Pintard, L., Willis, J.H., Hamill, D.R., Gonczy, P., Peter, M., and Bowerman, B. (2002). Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science 295, 1294–1298. Abstract Article
Li, J., Pauley, A.M., Myers, R.L., Shuang, R., Brashler, J.R., Yan, R., Buhl, A.E., Ruble, C., and Gurney, M.E. (2002). SEL-10 interacts with presenilin 1, facilitates its ubiquitination, and alters A-beta peptide production. J. Neurochem. 82, 1540–1548. Abstract Article
Liao, E.H., Hung, W., Abrams, B., and Zhen, M. (2004). An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature 430, 345–350. Abstract Article
Liu, J., Vasudevan, S., and Kipreos, E.T. (2004). CUL-2 and ZYG-11 promote meiotic anaphase II and the proper placement of the anterior–posterior axis in C. elegans. Development 131, 3513–3525. Abstract Article
Maeda, I., Kohara, Y., Yamamoto, M., and Sugimoto, A. (2001). Large-scale analysis of gene function in Caenorhabditis elegans by high- throughput RNAi. Curr. Biol. 11, 171–176. Abstract Article
Mathias, N., Johnson, S.L., Winey, M., Adams, A.E., Goetsch, L., Pringle, J.R., Byers, B., and Goebl, M.G. (1996). Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol. Cell Biol. 16, 6634–6643. Abstract
Mehta, N., Loria, P.M., and Hobert, O. (2004). A genetic screen for neurite outgrowth mutants in Caenorhabditis elegans reveals a new function for the F-box ubiquitin ligase component LIN-23. Genetics 166, 1253–1267. Abstract Article
Moore, R., and Boyd, L. (2004). Analysis of RING finger genes required for embryogenesis in C. elegans. Genesis 38, 1–12. Abstract Article
Nakata, K., Abrams, B., Grill, B., Goncharov, A., Huang, X., Chisholm, A.D., and Jin, Y. (2005). Regulation of a DLK-1 and p38 MAP Kinase Pathway by the Ubiquitin Ligase RPM-1 Is Required for Presynaptic Development. Cell 120, 407–420. Abstract Article
Nayak, S., Santiago, F.E., Jin, H., Lin, D., Schedl, T., and Kipreos, E.T. (2002). The Caenorhabditis elegans Skp1-related gene family: diverse functions in cell proliferation, morphogenesis, and meiosis. Curr. Biol. 12, 277–287. Abstract Article
Oberg, C., Li, J., Pauley, A., Wolf, E., Gurney, M., and Lendahl, U. (2001). The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 276, 35847–35853. Abstract Article
Ohta, T., and Fukuda, M. (2004). Ubiquitin and breast cancer. Oncogene 23, 2079–2088. Abstract Article
Ohta, T., Michel, J.J., Schottelius, A.J., and Xiong, Y. (1999). ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3, 535–541. Abstract Article
Ozkaynak, E., Finley, D., Solomon, M.J., and Varshavsky, A. (1987). The yeast ubiquitin genes: a family of natural gene fusions. EMBO J. 6, 1429–1439. Abstract
Page, A.P., MacNiven, K., and Hengartner, M.O. (1996). Cloning and biochemical characterization of the cyclophilin homologues from the free-living nematode Caenorhabditis elegans. Biochem. J. 317 (Pt 1), 179–185. Abstract
Pan, Z.Q., Kentsis, A., Dias, D.C., Yamoah, K., and Wu, K. (2004). Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23, 1985–1997. Abstract Article
Passmore, L.A., and Barford, D. (2004). Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem. J. 379, 513–525. Abstract Article
Peters, J.M. (2002). The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell 9, 931–943. Abstract Article
Petriv, O.I., Pilgrim, D.B., Rachubinski, R.A., and Titorenko, V.I. (2002). RNA interference of peroxisome-related genes in C. elegans: a new model for human peroxisomal disorders. Physiol. Genomics 10, 79–91. Abstract Article
Piano, F., Schetter, A.J., Mangone, M., Stein, L., and Kemphues, K.J. (2000). RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol. 10, 1619–1622. Abstract Article
Piano, F., Schetter, A.J., Morton, D.G., Gunsalus, K.C., Reinke, V., Kim, S.K., and Kemphues, K.J. (2002). Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr. Biol. 12, 1959–1964. Abstract Article
Pickart, C.M., and Cohen, R.E. (2004). Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5, 177–187. Abstract Article
Pintard, L., Kurz, T., Glaser, S., Willis, J.H., Peter, M., and Bowerman, B. (2003). Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr. Biol. 13, 911–921. Abstract Article
Pintard, L., Willis, J.H., Willems, A., Johnson, J.L., Srayko, M., Kurz, T., Glaser, S., Mains, P.E., Tyers, M., Bowerman, B., and Peter, M. (2003). The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425, 311–316. Abstract Article
Rappleye, C.A., Tagawa, A., Lyczak, R., Bowerman, B., and Aroian, R.V. (2002). The anaphase-promoting complex and separin are required for embryonic anterior-posterior axis formation. Dev. Cell 2, 195–206. Abstract Article
Rock, K.L., Gramm, C., Rothstein, L., Clark, K., Stein, R., Dick, L., Hwang, D., and Goldberg, A.L. (1994). Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771. Abstract Article
Salah, S.M., and Nasmyth, K. (2000). Destruction of the securin Pds1p occurs at the onset of anaphase during both meiotic divisions in yeast. Chromosoma 109, 27–34. Abstract Article
Sasagawa, Y., Urano, T., Kohara, Y., Takahashi, H., and Higashitani, A. (2003). Caenorhabditis elegans RBX1 is essential for meiosis, mitotic chromosomal condensation and segregation, and cytokinesis. Genes Cells 8, 857–872. Abstract Article
Schaefer, A.M., Hadwiger, G.D., and Nonet, M.L. (2000). rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron 26, 345–356. Abstract Article
Schlesinger, M.J., and Bond, U. (1987). Ubiquitin genes. Oxf. Surv. Eukaryot. Genes 4, 77–91. Abstract
Schneider, S.Q., and Bowerman, B. (2003). Cell polarity and the cytoskeleton in the Caenorhabditis elegans zygote. Annu. Rev. Genet. 37, 221–249. Abstract Article
Schnell, J.D., and Hicke, L. (2003). Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 278, 35857–35860. Abstract Article
Schulze, E., Altmann, M.E., Adham, I.M., Schulze, B., Frode, S., and Engel, W. (2003). The maintenance of neuromuscular function requires UBC-25 in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 305, 691–699. Abstract Article
Shakes, D.C., Sadler, P.L., Schumacher, J.M., Abdolrasulnia, M., and Golden, A. (2003). Developmental defects observed in hypomorphic anaphase-promoting complex mutants are linked to cell cycle abnormalities. Development 130, 1605–1620. Abstract Article
Shtiegman, K., and Yarden, Y. (2003). The role of ubiquitylation in signaling by growth factors: implications to cancer. Semin. Cancer Biol. 13, 29–40. Abstract Article
Simmer, F., Moorman, C., Van Der Linden, A.M., Kuijk, E., Van Den Berghe, P.V., Kamath, R., Fraser, A.G., Ahringer, J., and Plasterk, R.H. (2003). Genome-Wide RNAi of C. elegans Using the Hypersensitive rrf-3 Strain Reveals Novel Gene Functions. PLoS Biol. 1, E12. Abstract Article
Siomos, M.F., Badrinath, A., Pasierbek, P., Livingstone, D., White, J., Glotzer, M., and Nasmyth, K. (2001). Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr. Biol. 11, 1825–1835. Abstract Article
Slack, F.J., Basson, M., Liu, Z., Ambros, V., Horvitz, H.R., and Ruvkun, G. (2000). The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5, 659–669. Abstract Article
Sonneville, R., and Gonczy, P. (2004). Zyg-11 and cul-2 regulate progression through meiosis II and polarity establishment in C. elegans. Development 131, 3527–3543. Abstract Article
Takahashi, M., Iwasaki, H., Inoue, H., and Takahashi, K. (2002). Reverse genetic analysis of the Caenorhabditis elegans 26S proteasome subunits by RNA interference. Biol. Chem. 383, 1263–1266. Abstract Article
Thieringer, H., Moellers, B., Dodt, G., Kunau, W.H., and Driscoll, M. (2003). Modeling human peroxisome biogenesis disorders in the nematode Caenorhabditis elegans. J. Cell Sci. 116, 1797–1804. Abstract Article
Tijsterman, M., Ketting, R.F., Okihara, K.L., Sijen, T., and Plasterk, R.H. (2002). RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science 295, 694–697. Abstract Article
van den Heuvel, S. (2004). Protein degradation: CUL-3 and BTB–partners in proteolysis. Curr. Biol. 14, R59–R61. Abstract Article
Vodermaier, H.C. (2004). APC/C and SCF: controlling each other and the cell cycle. Curr. Biol. 14, R787–R796. Abstract Article
Wertz, I.E., O'Rourke, K.M., Zhang, Z., Dornan, D., Arnott, D., Deshaies, R.J., and Dixit, V.M. (2004). Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303, 1371–1374. Abstract Article
Wojcik, C., and DeMartino, G.N. (2003). Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 35, 579–589. Abstract Article
Wu, G., Hubbard, E.J., Kitajewski, J.K., and Greenwald, I. (1998). Evidence for functional and physical association between Caenorhabditis elegans SEL-10, a Cdc4p-related protein, and SEL-12 presenilin. Proc. Natl. Acad. Sci. USA 95, 15787–15791. Abstract Article
Wu, G., Lyapina, S., Das, I., Li, J., Gurney, M., Pauley, A., Chui, I., Deshaies, R.J., and Kitajewski, J. (2001). SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol. Cell Biol. 21, 7403–7415. Abstract Article
Wu, G., Xu, G., Schulman, B.A., Jeffrey, P.D., Harper, J.W., and Pavletich, N.P. (2003). Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF (beta-TrCP1) ubiquitin ligase. Mol. Cell 11, 1445–1456. Abstract Article
Xu, L., Wei, Y., Reboul, J., Vaglio, P., Shin, T.H., Vidal, M., Elledge, S.J., and Harper, J.W. (2003). BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425, 316–321. Abstract Article
Yamanaka, A., Yada, M., Imaki, H., Koga, M., Ohshima, Y., and Nakayama, K. (2002). Multiple Skp1-related proteins in Caenorhabditis elegans: diverse patterns of interaction with Cullins and F-box proteins. Curr. Biol. 12, 267–275. Abstract Article
Yanase, S., and Ishi, N. (1999). Cloning of the oxidative stress-responsive genes in Caenorhabditis elegans. J. Radiat. Res. (Tokyo) 40, 39–47. Abstract Article
Yeong, F.M. (2004). Anaphase-promoting complex in Caenorhabditis elegans. Mol. Cell Biol. 24, 2215–2225. Abstract Article
Yoon, C.H., Chang, C., Hopper, N.A., Lesa, G.M., and Sternberg, P.W. (2000). Requirements of multiple domains of SLI-1, a Caenorhabditis elegans homologue of c-Cbl, and an inhibitory tyrosine in LET-23 in regulating vulval differentiation. Mol. Biol. Cell 11, 4019–4031. Abstract
Yoon, C.H., Lee, J., Jongeward, G.D., and Sternberg, P.W. (1995). Similarity of sli-1, a regulator of vulval development in C. elegans, to the mammalian proto-oncogene c-cbl. Science 269, 1102–1105. Abstract
Zhen, M., Huang, X., Bamber, B., and Jin, Y. (2000). Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron 26, 331–343. Abstract Article
*Edited by Thomas Blumenthal. Last revised February 8, 2005. Published December 01, 2005. This chapter should be cited as: Kipreos, E.T. Ubiquitin-mediated pathways in C. elegans (December 01, 2005), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.36.1, http://www.wormbook.org.
§To whom correspondence should be addressed. E-mail: ekipreos@cb.uga.edu
 All WormBook content, except where otherwise noted, is licensed under a Creative Commons Attribution License.
All WormBook content, except where otherwise noted, is licensed under a Creative Commons Attribution License.